Research Projects
Summary: Our motivation for doing research is to move from correlation to causation in studies of host-microbe evolution. As oceans undergo major changes due to human activities (for example ocean warming and acidification), understanding how animals and plants adapt to a changing environment is now more than ever one of the biggest questions in marine biology. To predict future responses, we can explore the past and use geological events, which provide valuable insights into adaptive mechanisms.
The formation of the Isthmus of Panamá, for example, separated a previous, ancient ocean and all of its marine life into the Pacific Ocean and the Caribbean Sea. These two oceans have developed into very different habitats. Closely related populations that were separated by the Isthmus either went extinct or had to adapt to diverging environmental conditions. Therefore, the Isthmus system offers an ideal opportunity to explore drivers and processes of speciation, diversification, and adaptation through a convergent evolution framework.
Microorganisms, e.g., archaea, bacteria, viruses, or fungi, play a major role in host responses to environmental change because they can have significant effects on host health and fitness. In our team, we study the convergent evolution of a group of closely related hosts and their associated bacteria on both sides of the Isthmus of Panamá. We are comparing lucinid clam populations (Lucinidae) and their endosymbiotic bacterial chemosymbionts (Candidatus Thiodiazotropha) at the genomic and transcriptomic level across the Isthmus of Panamá. In doing so, we can learn how a dramatic allopatric speciation event affected host-microbe associations from genotype to phenotype.
We are convinced that microbial partners are the key for understanding niche partitioning in highly diverse habitats, host evolution, and ecosystem functioning. We have three major research axes:
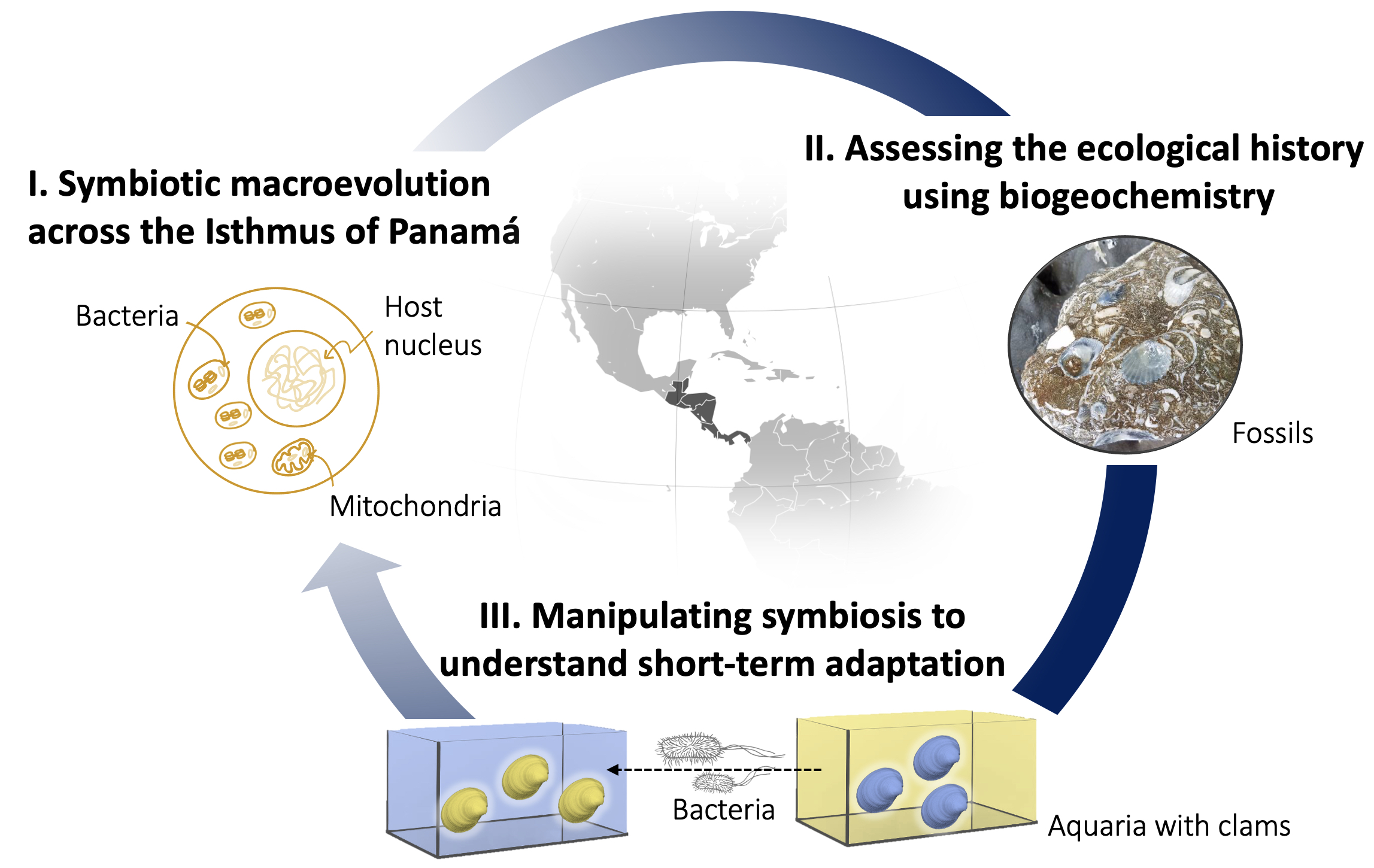
I. What are the rates of molecular divergence and signatures of past adaptation in host and symbiont genomes?
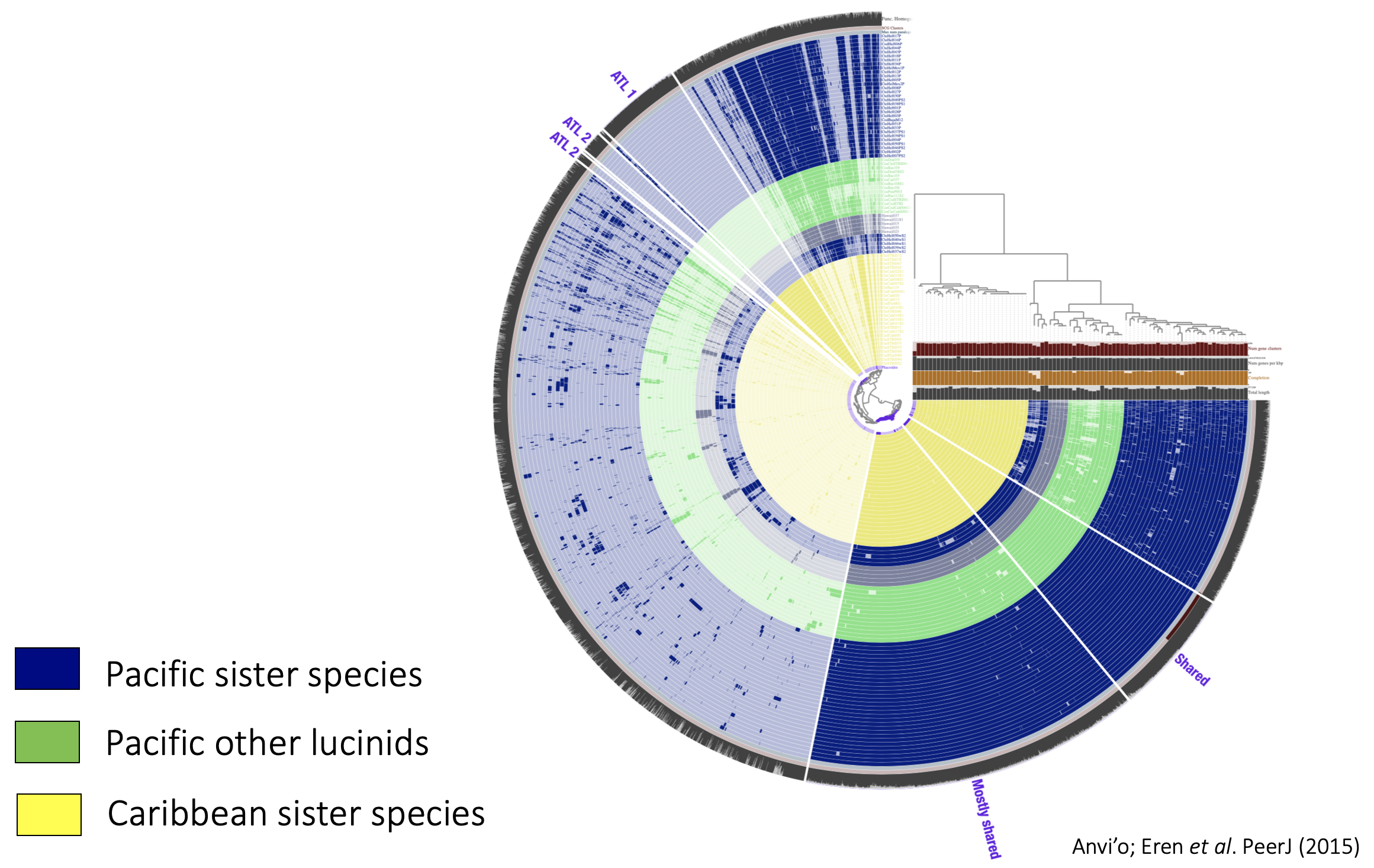
Based on sequencing data, we are reconstructing the molecular divergence of eukaryotic hosts and their microbial partners after they were separated by the Isthmus of Panamá, and we are characterizing genes that facilitated survival in different habitats. Using long-read sequencing, we are able to achieve an unprecedented level of resolution and reconstruct entire gene families in both hosts and microbes. Shared gene families allow us to investigate what the two partners in the symbiosis have been doing and at what speed are they doing it? We are comparing the rates of molecular divergence among the three genomes (host nuclear, mitochondrial and bacterial symbiont). Complex metazoan genomes vs. streamlined bacterial genomes. Who evolves faster? Is it the host or the symbiont that adapts? And how does the interplay between bacteria and host facilitate adaptation of the meta-organism to new conditions?
II. What were the ecological conditions in the past and how did this affect host-microbe interactions?
Here, we are moving beyond genetic markers and using the fossil record to understand the biogeochemical past of a symbiosis. We are looking into the past to understand the future. Fossils allow us to calibrate rates of evolution and study past host-microbe interactions. We are particularly interested in the nitrogen cycle.

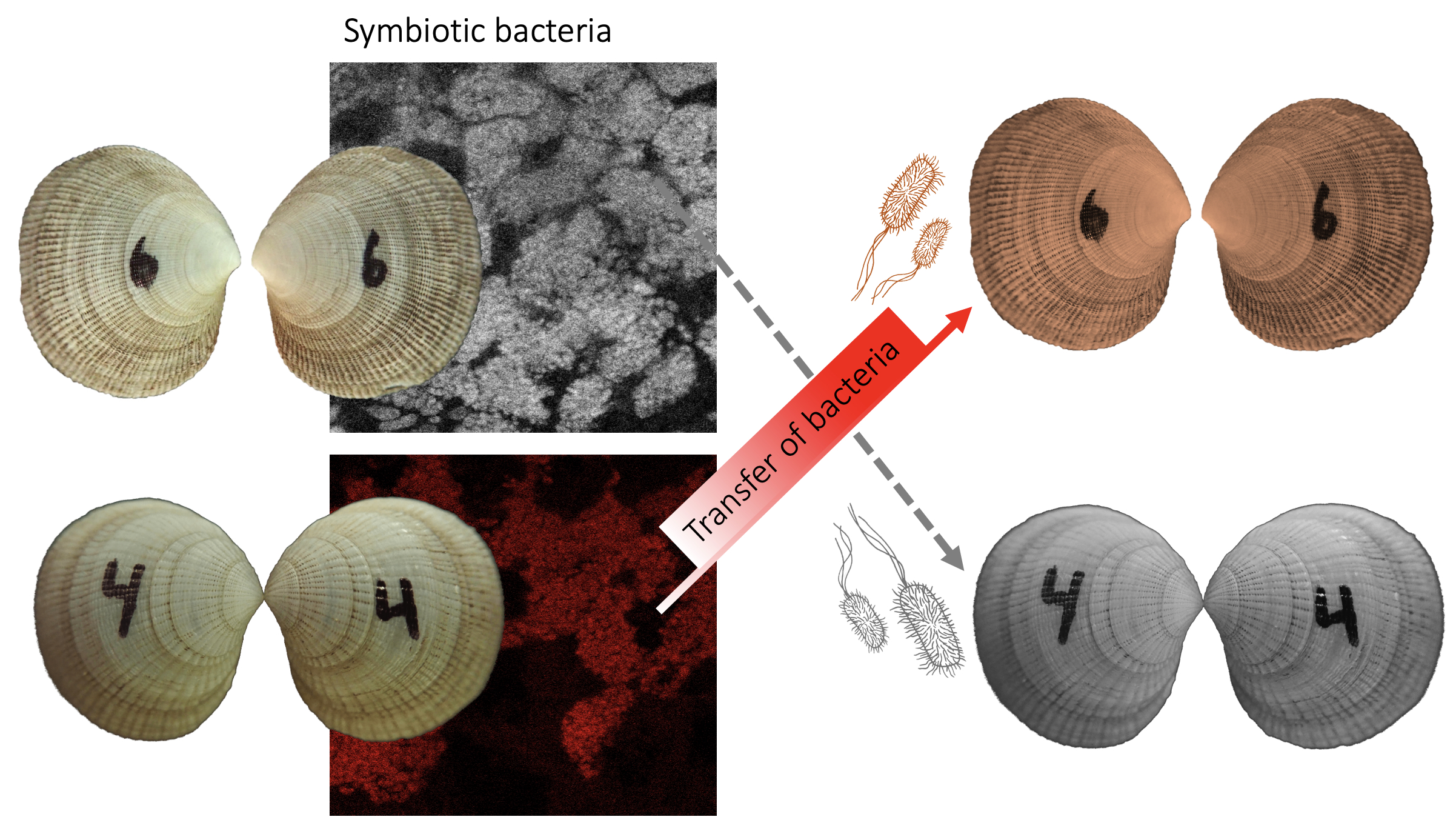
III. How quickly can an animal-microbial system adapt to new conditions?
For this project, we are manipulating the symbiosis in the lab and creating new host-symbiont combinations. These will be exposed to different environments to induce short-term adaptation. The combination of symbiont exchange and common garden experiments lets us answer key questions in symbiosis research and give insight into the mechanisms of host-symbiont interaction. Using cutting-edge imaging techniques at the Max Planck Institute, we are able to visualize expressed genes and exchanged metabolites in real time and quantify how they affect host fitness and survival.